Concurrent loss of the Y chromosome in cancer and T cells impacts outcome
Chen, X., Shen, Y., Choi, S. et al. Nature 642, 1041–1050 (2025). First author. https://doi.org/10.1038/s41586-025-09071-2 a>
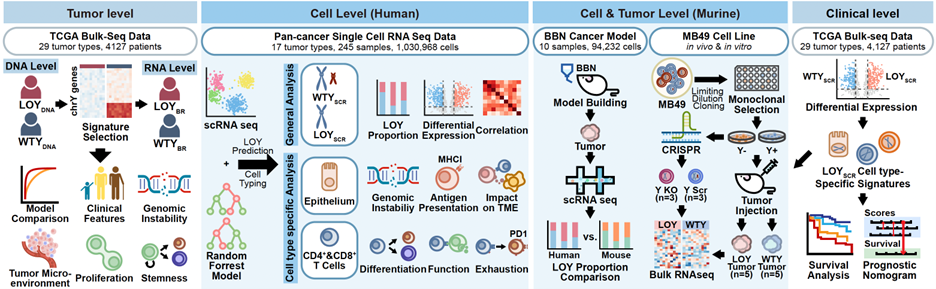
In our 2025 Nature study (first authored by me), we demonstrated that age-related loss of the Y chromosome (LOY)—which affects up to 40% of elderly men—occurs not only in tumour cells but also spreads to infiltrating T cells; this “double-LOY” condition cripples anti-tumour immunity and predicts the worst survival across 4,000 male cancer samples and more than one million single-cell profiles. Mechanistically, losing key Y-linked genes (KDM5D, UTY, DDX3Y) weakens tumour antigen presentation, while LOY T cells become exhausted (↓ GZMB/PRF1, ↑ PD-1/CTLA-4), offering an explanation for men’s higher cancer burden. These findings argue for routine LOY testing in male-predominant cancers, careful exclusion of LOY T cells from cell therapies, and development of treatments that restore or block LOY-driven immune collapse.
Nature News & Views (4 June 2025) spotlighted our Nature paper revealing Y-chromosome loss (LOY) in both tumour cells and infiltrating T cells. Commentators Nicholas McGranahan (University College London) and Rahul Roychoudhuri (University of Cambridge) contend that this coordinated LOY weakens antitumour immunity, enables escape, and predicts poorer survival. They urge clinicians to assess LOY in both compartments for prognosis, use Y-status to guide immuno- and cell-therapy choices, and investigate how Y-deficient tumours trigger LOY in nearby immune cells to discover new therapeutic targets.